Basic information
- Phenotype
- HALLMARK_TGF_BETA_SIGNALING
- Description
- Enrichment score representing TGFbeta signaling. The score was calculated using ssGSEA on the RNA data (Z-score transformed coding genes with expression in at least 50% of the samples in a cohort).
- Source
- http://www.gsea-msigdb.org/gsea/msigdb/cards/HALLMARK_TGF_BETA_SIGNALING.html
- Method
- Single sample gene set enrichment analysis (ssGSEA) was performed for each cancer type using gene-wise Z-scores of the RNA expression data (RSEM) for the MSigDB Hallmark gene sets v7.0 (PMID: 26771021) via the ssGSEA2.0 R package (PMID: 30563849). RNA data were filtered to coding genes with < 50% 0 expression. (Parameters: sample.norm.type="rank", weight=0.75, statistic="area.under.RES", nperm=1000, min.overlap=10). Pathway activity scores are normalized enrichment scores from ssGSEA.
- Genes
-
- APC
- ARID4B
- BCAR3
- BMP2
- BMPR1A
- BMPR2
- CDH1
- CDK9
- CDKN1C
- CTNNB1
- ENG
- FKBP1A
- FNTA
- FURIN
- HDAC1
- HIPK2
- ID1
- ID2
- ID3
- IFNGR2
- JUNB
- KLF10
- LEFTY2
- LTBP2
- MAP3K7
- NCOR2
- NOG
- PMEPA1
- PPM1A
- PPP1CA
- PPP1R15A
- RAB31
- RHOA
- SERPINE1
- SKI
- SKIL
- SLC20A1
- SMAD1
- SMAD3
- SMAD6
- SMAD7
- SMURF1
- SMURF2
- SPTBN1
- TGFB1
- TGFBR1
- TGIF1
- THBS1
- TJP1
- TRIM33
- UBE2D3
- WWTR1
- XIAP
- More...
Gene association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in bar plots. The scatter plot highlights significant associations diven by protein rather than mRNA abundance.
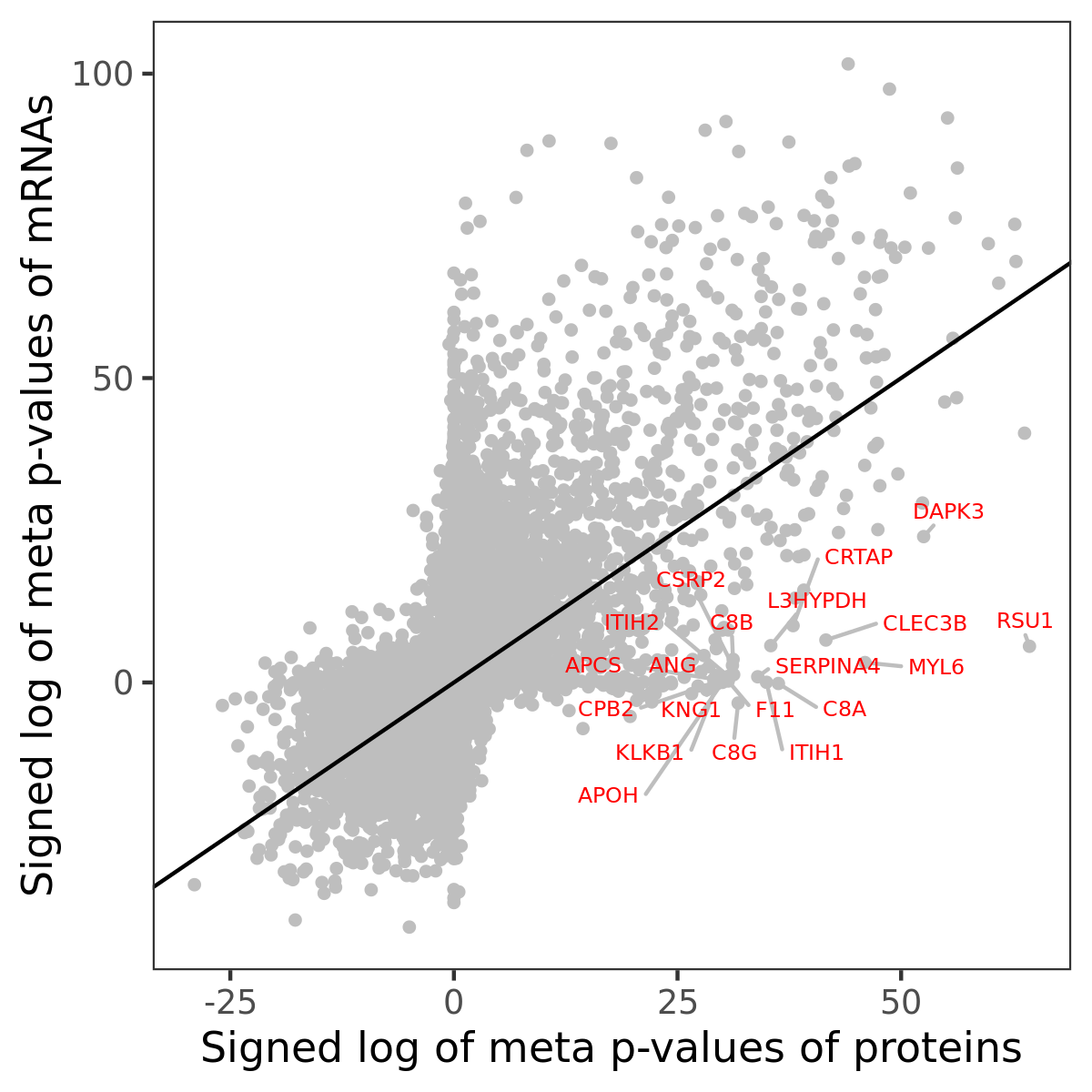
Association of protein abundance of genes
| Signed p-values | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC |
Gene set enrichment analysis
Submit genes and the common logarithm of the p-values of their association with HALLMARK_TGF_BETA_SIGNALING to WebGestalt.
Phosphosite association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in the bar plot. The scatter plot highlights significant associations diven by phosphorylation rather than protein abundance.
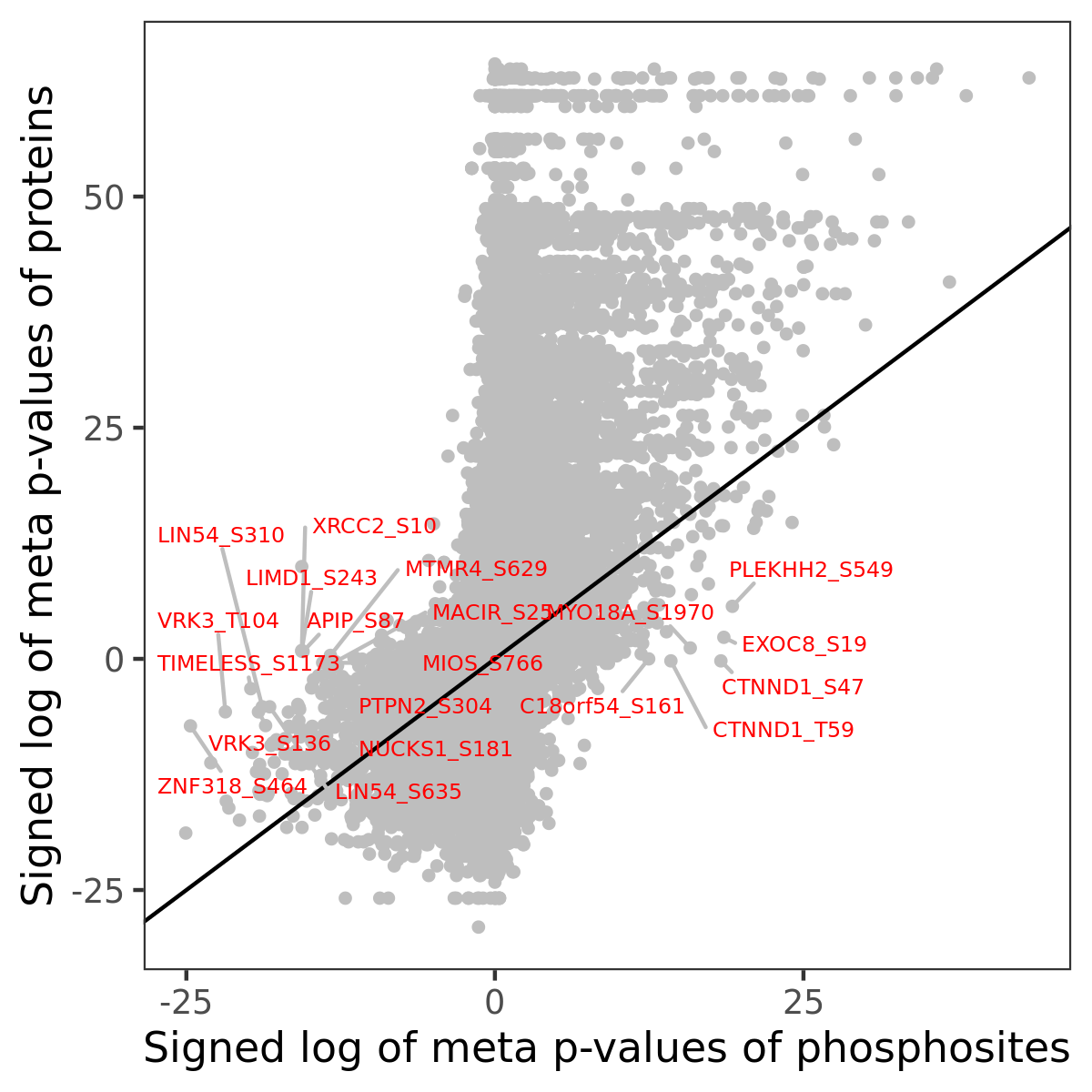
| Signed p-values | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Protein | Site | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC |
Gene set enrichment analysis
Submit phosphorylation sites and the common logarithm of the p-values of their association with HALLMARK_TGF_BETA_SIGNALING to WebGestalt.