Basic information
- Phenotype
- xcell: Common myeloid progenitor
- Description
- Enrichment score inferring the proportion of common myeloid progenitor cells in the tumor derived from RNA data using the xCell tool.
- Source
- https://genomebiology.biomedcentral.com/articles/10.1186/s13059-017-1349-1
- Method
- The R package immunedeconv (V2.0.4) (PMID: 31510660) was used to perform immune cell deconvolution using RNA expression data (TPM).
- Genes
-
- AFF2
- ALOX15
- ALX4
- ANKRD34C
- ANXA3
- ARHGAP33
- ATP8B3
- ATP8B4
- ATXN7L1
- AZU1
- BAHCC1
- BFSP2
- BRSK2
- BYSL
- C1QB
- C21orf62
- CACNA1E
- CACNG3
- CADM3
- CAMK2A
- CCDC121
- CCL18
- CCL23
- CCNB2
- CCNC
- CD1E
- CD33
- CD63
- CDSN
- CDT1
- CENPJ
- CENPN
- CHAF1A
- CLC
- CLIC1
- CLPP
- CNGA3
- CNTN6
- COG7
- COL14A1
- COMMD4
- CPA3
- CPB1
- CPN1
- CRHBP
- CRTAC1
- CRYGA
- CRYGD
- CSF2RB
- CTSG
- CYP3A43
- CYSLTR2
- DAB1
- DAZAP1
- DEFA6
- DHX32
- DLEC1
- DNAI1
- DNASE2B
- DPEP3
- EDN3
- EHMT2
- ELANE
- EPX
- ERLIN1
- EXD3
- EYA3
- FAIM2
- FAM111A
- FAM76A
- FANCG
- FBXL4
- FHL5
- FLT3
- FMO2
- FOXB1
- FRMPD4
- G6PC
- GALR1
- GDF5
- GDF9
- GLYAT
- GNAT1
- GNMT
- GPD1
- GPR135
- GPR27
- GRIA4
- GRM2
- GUCA2B
- HDAC6
- HDC
- HMGXB3
- HPGDS
- HRH4
- HSF4
- IDUA
- IGLL1
- IL13
- IL5RA
- IMPG1
- IRF2BP1
- IRS4
- ITIH3
- ITIH4
- JRK
- KCNJ10
- KCNN1
- KLHL11
- KLHL9
- L2HGDH
- LCT
- LIN28A
- LIPF
- LONP1
- LPAR4
- LRRC19
- LRRC3
- LTC4S
- LYL1
- MADCAM1
- MAP2K5
- MAPK14
- MEA1
- MED18
- MGA
- MKI67
- MPL
- MPO
- MS4A2
- MS4A3
- MTHFSD
- MYF5
- MYH11
- MYLPF
- MYO9A
- NAALADL1
- NDOR1
- NDUFB4
- NEU3
- NRIP2
- NSMAF
- NTN1
- OXT
- P2RY4
- PAX1
- PAX3
- PDIA2
- PDX1
- PGM1
- PGR
- PHF7
- PIKFYVE
- PLA2G6
- POLA2
- POLE
- PRG2
- PRG3
- PRKG2
- PRODH2
- PRTN3
- Array
- PTPRS
- PTPRT
- PYY
- RAB40A
- RASL10A
- RASL12
- RCVRN
- RERGL
- REV1
- RHO
- RHOT1
- RNASE2
- RNASE3
- RORB
- RREB1
- RUNX1
- RUVBL2
- S100PBP
- SCAMP1
- SCN1A
- SERPINB10
- SERPINI2
- SETD5
- SIM1
- SLC13A2
- SLC18A2
- SLC4A5
- SLC5A2
- SMARCAL1
- SMC5
- SNAPC4
- SNW1
- SNX5
- SPAG5
- SPAG8
- SPATA7
- SPN
- STAR
- TAB1
- TAF6L
- TBC1D16
- TFCP2
- TGM5
- TIMM50
- TNPO3
- TNR
- TOLLIP
- TOP2B
- TOR1A
- TP73
- TPSAB1
- TRAIP
- TRIM27
- TRPC3
- TSGA10
- TUBA1B
- TYR
- UBQLN3
- UNG
- USP19
- USP48
- VN1R1
- VPREB1
- VPS54
- XPNPEP3
- XPO1
- ZBBX
- ZKSCAN3
- ZKSCAN4
- ZKSCAN5
- ZMAT5
- ZMYM4
- ZNF174
- ZNF197
- ZNF221
- ZNF324B
- ZNF428
- ZNF471
- ZNF629
- ZNF701
- ZNF747
- ZNF768
- ZNF780B
- More...
Gene association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in bar plots. The scatter plot highlights significant associations diven by protein rather than mRNA abundance.

Association of protein abundance of genes
Signed p-values | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Gene | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC | |
| No matching records found | ||||||||||||
Gene set enrichment analysis
Submit genes and the common logarithm of the p-values of their association with xcell: Common myeloid progenitor to WebGestalt.
Phosphosite association
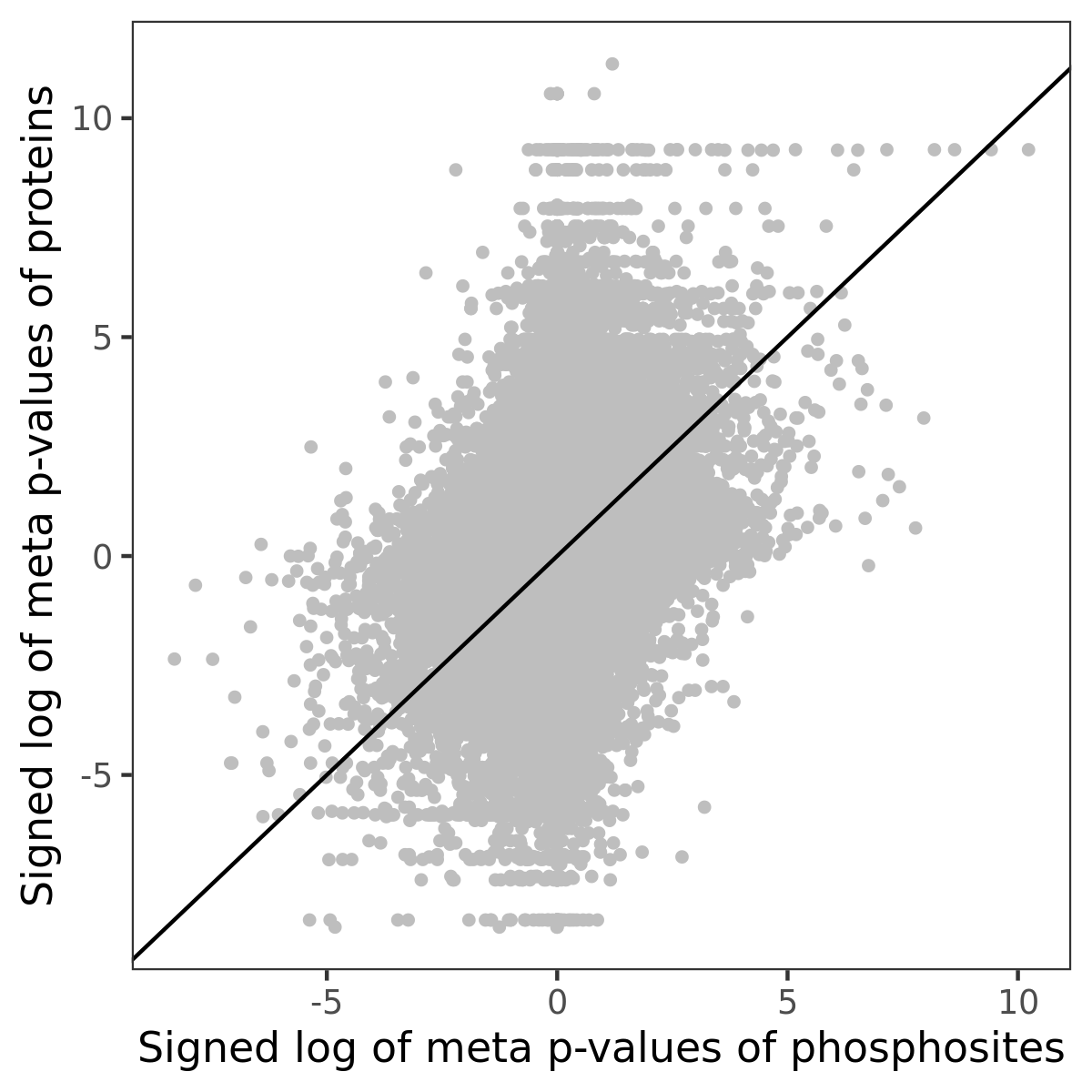
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in the bar plot. The scatter plot highlights significant associations diven by phosphorylation rather than protein abundance.
Signed p-values | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Gene | Protein | Site | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC |
| No matching records found | |||||||||||||
Gene set enrichment analysis
Submit phosphorylation sites and the common logarithm of the p-values of their association with xcell: Common myeloid progenitor to WebGestalt.