Basic information
- Phenotype
- HALLMARK_SPERMATOGENESIS
- Description
- Enrichment score representing genes involved in the process of spermatogenesis. The score was calculated using ssGSEA on the RNA data (Z-score transformed coding genes with expression in at least 50% of the samples in a cohort).
- Source
- http://www.gsea-msigdb.org/gsea/msigdb/cards/HALLMARK_SPERMATOGENESIS.html
- Method
- Single sample gene set enrichment analysis (ssGSEA) was performed for each cancer type using gene-wise Z-scores of the RNA expression data (RSEM) for the MSigDB Hallmark gene sets v7.0 (PMID: 26771021) via the ssGSEA2.0 R package (PMID: 30563849). RNA data were filtered to coding genes with < 50% 0 expression. (Parameters: sample.norm.type="rank", weight=0.75, statistic="area.under.RES", nperm=1000, min.overlap=10). Pathway activity scores are normalized enrichment scores from ssGSEA.
- Genes
-
- ACRBP
- ACRV1
- ACTL7B
- ADAD1
- ADAM2
- ADCYAP1
- AGFG1
- AKAP4
- ALOX15
- ARL4A
- ART3
- AURKA
- BRAF
- BUB1
- CAMK4
- CCNA1
- CCNB2
- CCT6B
- CDK1
- CDKN3
- CFTR
- CHFR
- CHRM4
- CLGN
- CLPB
- CLVS1
- CNIH2
- COIL
- CRISP2
- CSNK2A2
- CST8
- DBF4
- DCC
- DDX25
- DDX4
- DMC1
- DMRT1
- DNAJB8
- DPEP3
- ELOVL3
- EZH2
- GAD1
- GAPDHS
- GFI1
- GMCL1
- GPR182
- GRM8
- GSG1
- GSTM3
- H1-6
- HBZ
- HOXB1
- HSPA1L
- HSPA2
- HSPA4L
- HTR5A
- IDE
- IFT88
- IL12RB2
- IL13RA2
- IP6K1
- JAM3
- KIF2C
- LDHC
- LPIN1
- MAP7
- MAST2
- MEP1B
- MLF1
- MLLT10
- MTNR1A
- MTOR
- NAA11
- NCAPH
- NEFH
- NEK2
- NF2
- NOS1
- NPHP1
- NPY5R
- OAZ3
- ODF1
- PACRG
- PAPOLB
- PARP2
- PCSK1N
- PCSK4
- PDHA2
- PEBP1
- PGK2
- PGS1
- PHF7
- PHKG2
- PIAS2
- POMC
- PRKAR2A
- PRM2
- PSMG1
- RAD17
- RFC4
- RPL39L
- SCG3
- SCG5
- SEPTIN4
- SHE
- SIRT1
- SLC12A2
- SLC2A5
- SNAP91
- SPATA6
- STAM2
- STRBP
- SYCP1
- TALDO1
- TCP11
- TEKT2
- THEG
- TKTL1
- TLE4
- TNNI3
- TNP1
- TNP2
- TOPBP1
- TSN
- TSSK2
- TTK
- TUBA3C
- TULP2
- VDAC3
- YBX2
- ZC2HC1C
- ZC3H14
- ZNRF4
- ZPBP
- More...
Gene association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in bar plots. The scatter plot highlights significant associations diven by protein rather than mRNA abundance.
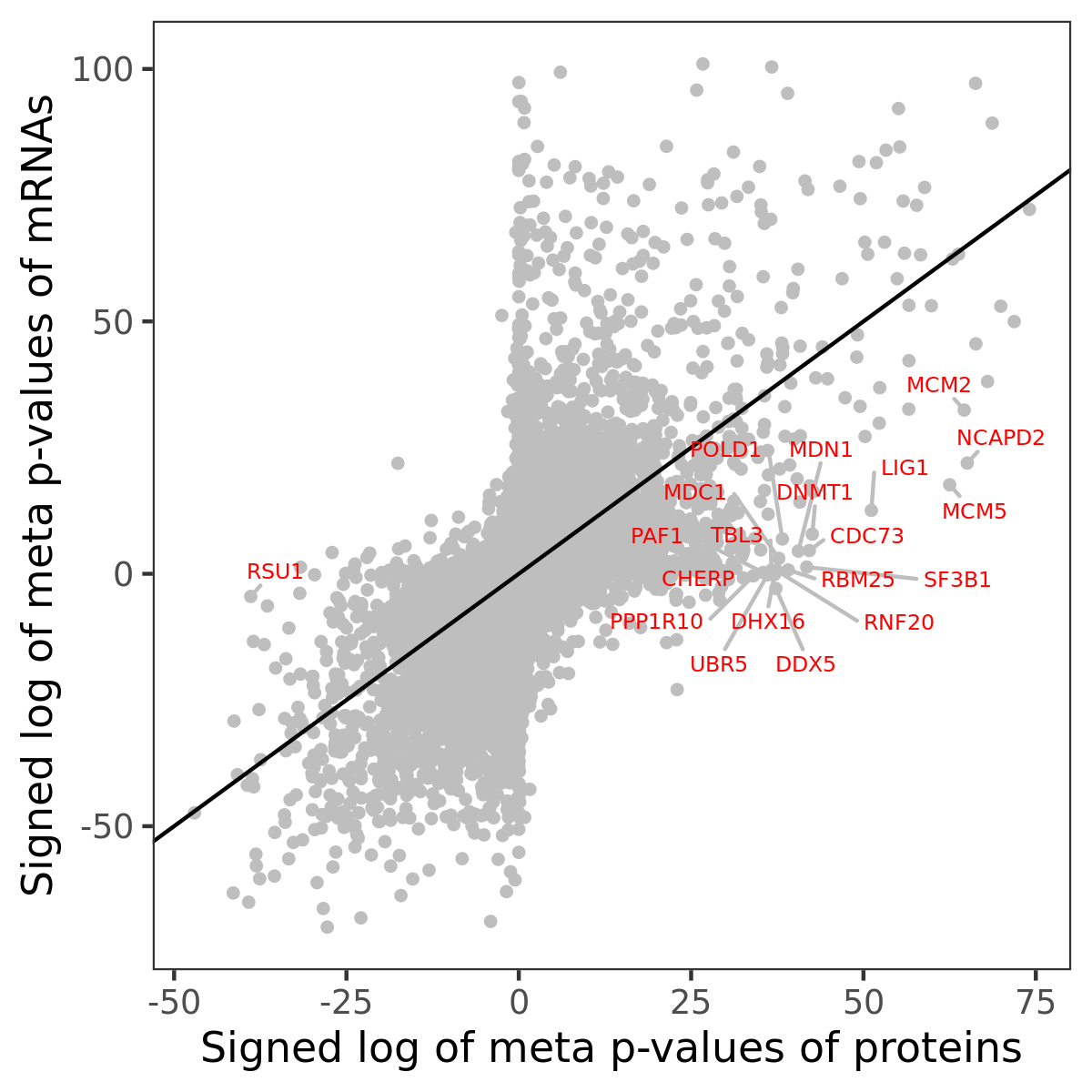
Association of protein abundance of genes
| Signed p-values | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC |
Gene set enrichment analysis
Submit genes and the common logarithm of the p-values of their association with HALLMARK_SPERMATOGENESIS to WebGestalt.
Phosphosite association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in the bar plot. The scatter plot highlights significant associations diven by phosphorylation rather than protein abundance.
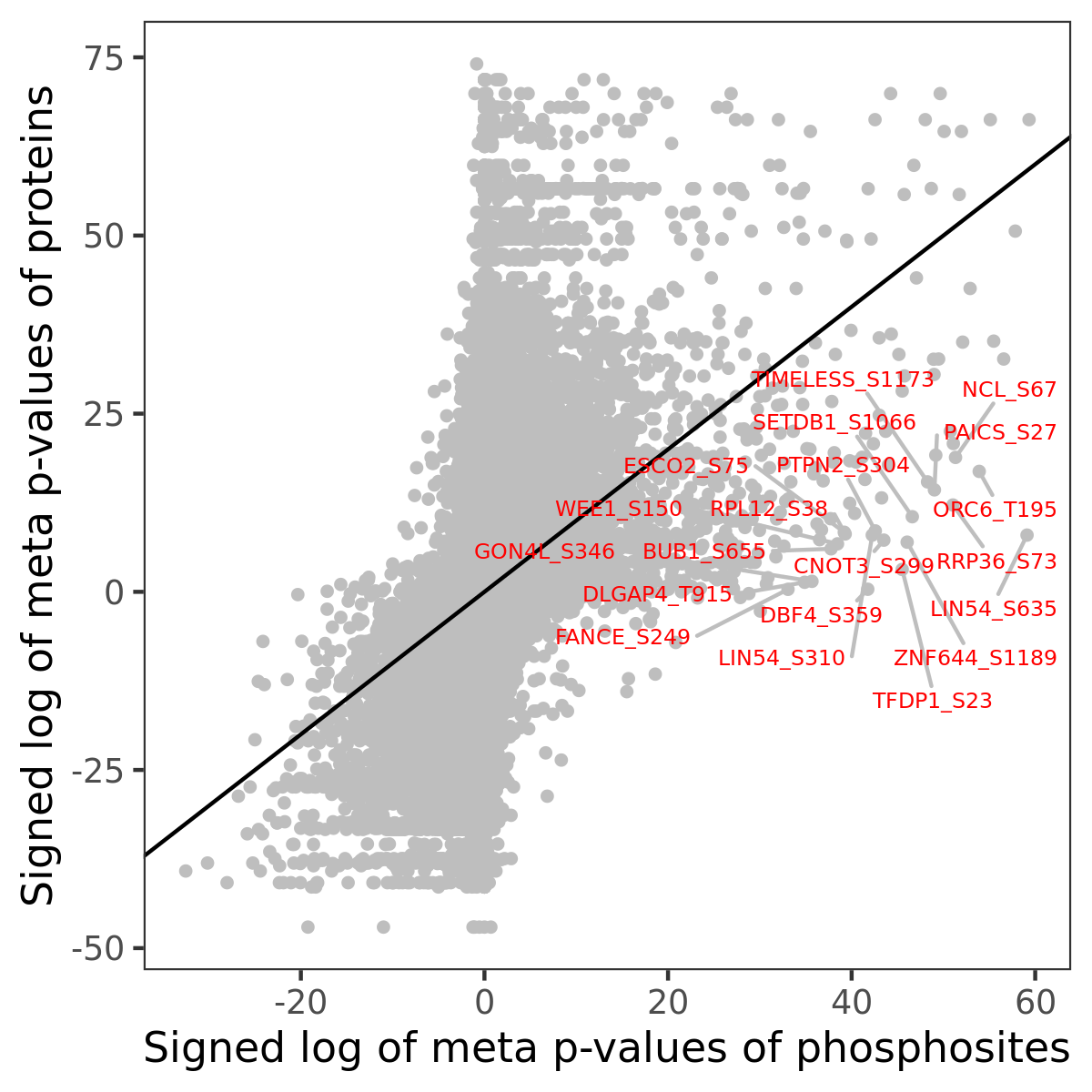
| Signed p-values | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Protein | Site | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC |
Gene set enrichment analysis
Submit phosphorylation sites and the common logarithm of the p-values of their association with HALLMARK_SPERMATOGENESIS to WebGestalt.