Basic information
- Phenotype
- xcell: Macrophage
- Description
- Enrichment score inferring the proportion of macrophages in the tumor derived from RNA data using the xCell tool.
- Source
- https://genomebiology.biomedcentral.com/articles/10.1186/s13059-017-1349-1
- Method
- The R package immunedeconv (V2.0.4) (PMID: 31510660) was used to perform immune cell deconvolution using RNA expression data (TPM).
- Genes
-
- ACADVL
- ACP2
- ACTR10
- ADAMDEC1
- ADCK2
- ADCY3
- AGPS
- ALCAM
- ARHGEF11
- ARL8B
- ARPC4
- ARSB
- ATOX1
- ATP2C1
- ATP6AP2
- ATP6V0A1
- ATP6V0C
- ATP6V0E1
- ATP6V1A
- ATP6V1C1
- ATP6V1D
- ATP6V1E1
- ATP6V1F
- ATP6V1H
- BAG3
- BAIAP2
- BCAP31
- BPI
- BTBD1
- C12orf4
- C12orf49
- C1QA
- C7orf25
- CCDC88A
- CCL1
- CCL18
- CCL22
- CCL7
- CCL8
- CCR1
- CD163
- CD164
- CD48
- CD63
- CD84
- CD9
- CEPT1
- CETN2
- CHD9
- CHIT1
- CIAO1
- CIR1
- CLCN7
- CLEC5A
- CLIP1
- CMKLR1
- CNIH4
- COMMD8
- COMMD9
- COX15
- COX5A
- COX5B
- COX7B
- COX8A
- CPNE6
- CRYBB1
- CSF1
- CXCL9
- CYBA
- CYBB
- CYP19A1
- DBI
- DERA
- DNAJC13
- DNASE2B
- ECHS1
- EFR3A
- ELOVL1
- EMILIN1
- ERP29
- FCER1G
- FDX1
- FEZ2
- FGR
- FKBP15
- FOLR2
- FPR3
- FTL
- G6PC3
- GLB1
- GLRX2
- GRB2
- GSTO1
- GUF1
- HADHB
- HAMP
- HCCS
- HEXA
- HEXB
- HIGD2A
- HK3
- HMGCL
- HS3ST2
- HSD17B12
- HTT
- IBTK
- IGSF6
- IL17RA
- ITGAX
- ITGB1BP1
- KCMF1
- KCNJ1
- KCNJ5
- KCNMB1
- KCTD5
- KIFC3
- KLHL12
- LAIR1
- LAMP1
- LDHAL6B
- LILRB4
- LILRB5
- LONP1
- LONRF3
- LY86
- M6PR
- MAPK13
- MAPKAP1
- MARCO
- MDH1
- MGST3
- MLX
- MMP19
- MMP8
- MRPL40
- MRS2
- MS4A4A
- MS4A6A
- MSR1
- MTHFR
- MTMR14
- MUL1
- MYOZ1
- NCAPH
- NDUFA8
- NDUFAF1
- NDUFB3
- NDUFB6
- NDUFS3
- NDUFS6
- NDUFS8
- NOP10
- NPTN
- NRBP1
- NSMAF
- NSUN3
- NUBP1
- NUDT9
- NUMB
- ORMDL2
- P2RX7
- PANK3
- PCDHB11
- PCMT1
- PDCD6IP
- PEX14
- PEX19
- PMFBP1
- PPCS
- PRDX1
- PRDX3
- PSMD10
- PSME1
- PTPN12
- PTPRA
- QDPR
- RAB1A
- RAB5C
- RABGGTA
- RAC1
- RALA
- RB1
- RENBP
- RTN4
- S1PR2
- SCAMP2
- SDHB
- SDHD
- SDS
- SETD3
- SH3GLB1
- SIGLEC7
- SIGLEC9
- SLAMF8
- SLC11A1
- SLC1A2
- SLC25A24
- SLC30A5
- SLC31A1
- SLC38A7
- SLC6A12
- SNUPN
- SNX2
- SNX3
- SNX4
- SPG21
- SPIN1
- SRC
- STAB1
- STAM2
- STX18
- STX4
- STYXL1
- SUMO3
- TBC1D16
- TCEAL4
- TGOLN2
- TM2D1
- TMBIM4
- TMEM115
- TMEM126B
- TMEM127
- TMEM147
- TMEM33
- TMEM70
- TMEM9B
- TNFRSF12A
- TNPO1
- TPP1
- TRAF3
- TRAPPC2L
- TRAPPC3
- TREM2
- TSPO
- TTLL4
- TULP4
- TYROBP
- UBE2D4
- UQCR10
- UQCR11
- UQCRC2
- USF2
- VAMP8
- VPS53
- VSIG4
- VTI1B
- WDR11
- YIF1B
- ZDHHC24
- ZDHHC3
- ZMPSTE24
- More...
Gene association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in bar plots. The scatter plot highlights significant associations diven by protein rather than mRNA abundance.
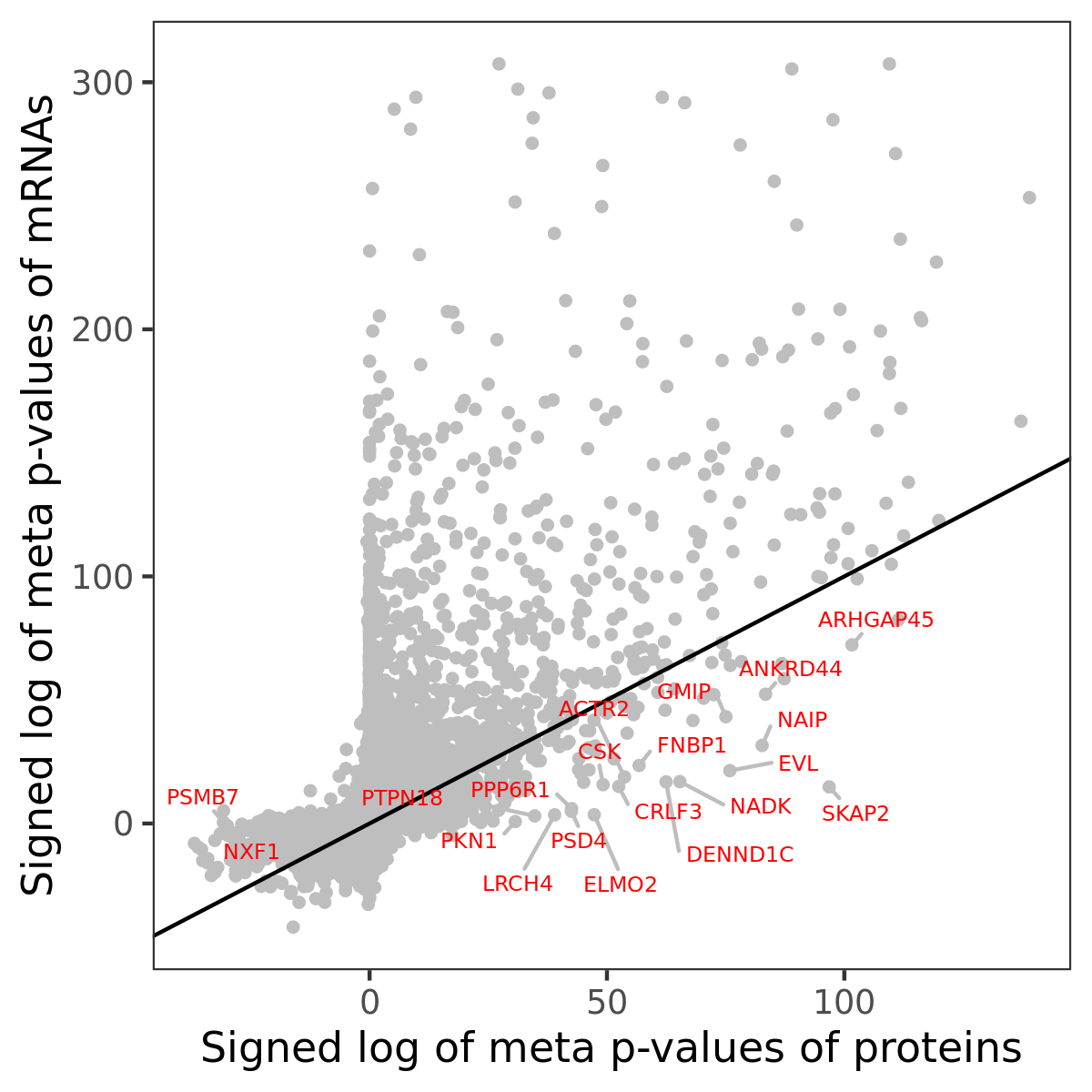
Association of protein abundance of genes
Signed p-values | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Gene | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC | |
| No matching records found | ||||||||||||
Gene set enrichment analysis
Submit genes and the common logarithm of the p-values of their association with xcell: Macrophage to WebGestalt.
Phosphosite association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in the bar plot. The scatter plot highlights significant associations diven by phosphorylation rather than protein abundance.
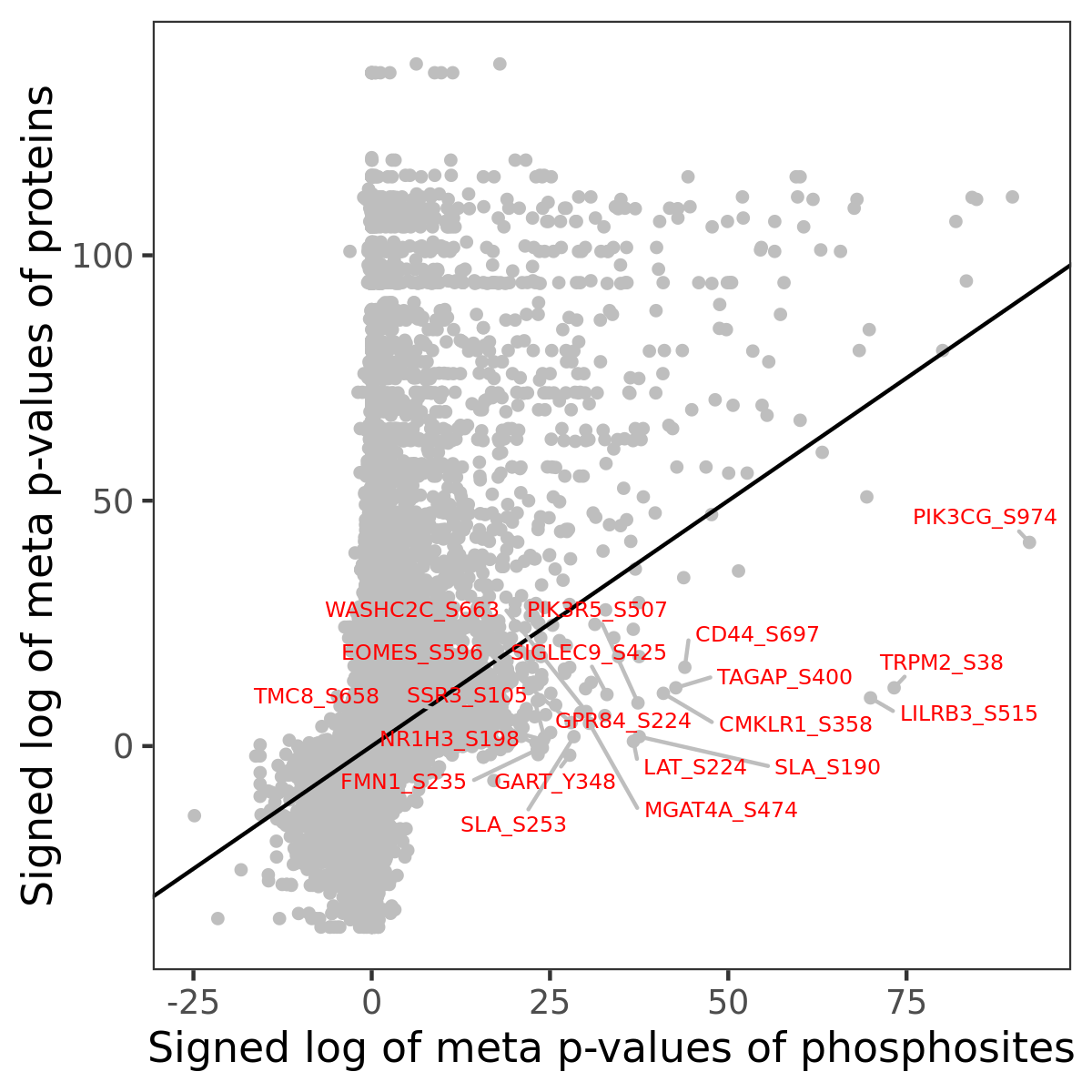
Signed p-values | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Gene | Protein | Site | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC |
| No matching records found | |||||||||||||
Gene set enrichment analysis
Submit phosphorylation sites and the common logarithm of the p-values of their association with xcell: Macrophage to WebGestalt.