Basic information
- Phenotype
- xcell: Mast cell
- Description
- Enrichment score inferring the proportion of mast cells in the tumor derived from RNA data using the xCell tool.
- Source
- https://genomebiology.biomedcentral.com/articles/10.1186/s13059-017-1349-1
- Method
- The R package immunedeconv (V2.0.4) (PMID: 31510660) was used to perform immune cell deconvolution using RNA expression data (TPM).
- Genes
-
- AGGF1
- AGPS
- AMHR2
- ANXA1
- ANXA11
- ARHGEF12
- ATP6V1C1
- ATXN10
- ATXN2L
- BAHD1
- BAIAP2
- BET1L
- BMPR1A
- BTBD7
- BTK
- C3AR1
- C8G
- CASP10
- CD22
- CD33
- CD84
- CDC16
- CENPJ
- CHD9
- CMA1
- CPA3
- CPSF1
- CTDSP1
- CTR9
- CTSG
- DCLRE1B
- DEPDC5
- DIAPH1
- DNAJC13
- DNAJC28
- DR1
- ESYT1
- FAM120A
- FBXO11
- FBXO38
- GATA1
- HDC
- HIBCH
- HPGDS
- HRH4
- HSPH1
- IFT27
- IL5
- IL5RA
- ITGA2B
- KCNJ5
- KLRG1
- KRT1
- LAX1
- LCP2
- LRIG2
- LTC4S
- LYL1
- MAGT1
- MAP3K7
- MBOAT7
- MRPS28
- MS4A2
- MTR
- NBAS
- NDST2
- NTRK1
- OSBP
- OSBPL9
- P2RX1
- PABPC4
- PAK2
- PDE4A
- PIK3R2
- POLR2A
- PPP3R1
- PRG2
- PRKAR1A
- PTGDR
- PTGER3
- RAD23B
- REM1
- RENBP
- RGS11
- RGS13
- RNF103
- RXRB
- SERINC3
- SIGLEC6
- SIGLEC7
- SIGLEC8
- SLC18A2
- SNRNP70
- SNUPN
- SNX5
- SOS2
- SPAG8
- STAM
- STAP1
- SYPL1
- TADA2A
- TAL1
- TEC
- THRAP3
- TIPRL
- TPSAB1
- TRIM32
- TTC17
- U2AF2
- UPF2
- USP10
- USP48
- WDR46
- ZBTB1
- ZBTB7A
- ZKSCAN3
- ZMYM1
- ZMYM4
- ZNF212
- ZNF264
- ZNF426
- ZNF471
- ZNF549
- More...
Gene association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in bar plots. The scatter plot highlights significant associations diven by protein rather than mRNA abundance.
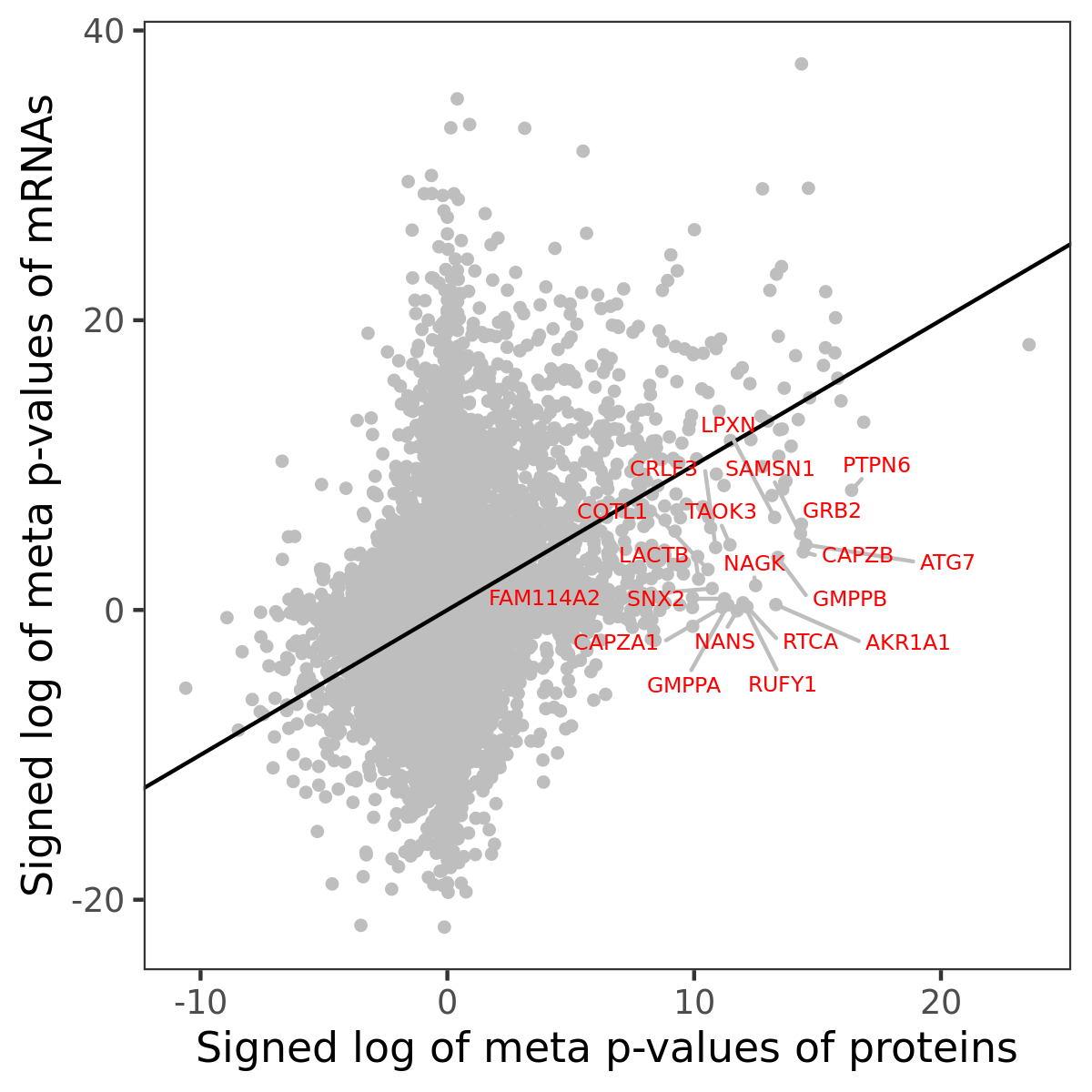
Association of protein abundance of genes
| Signed p-values | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC |
Gene set enrichment analysis
Submit genes and the common logarithm of the p-values of their association with xcell: Mast cell to WebGestalt.
Phosphosite association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in the bar plot. The scatter plot highlights significant associations diven by phosphorylation rather than protein abundance.
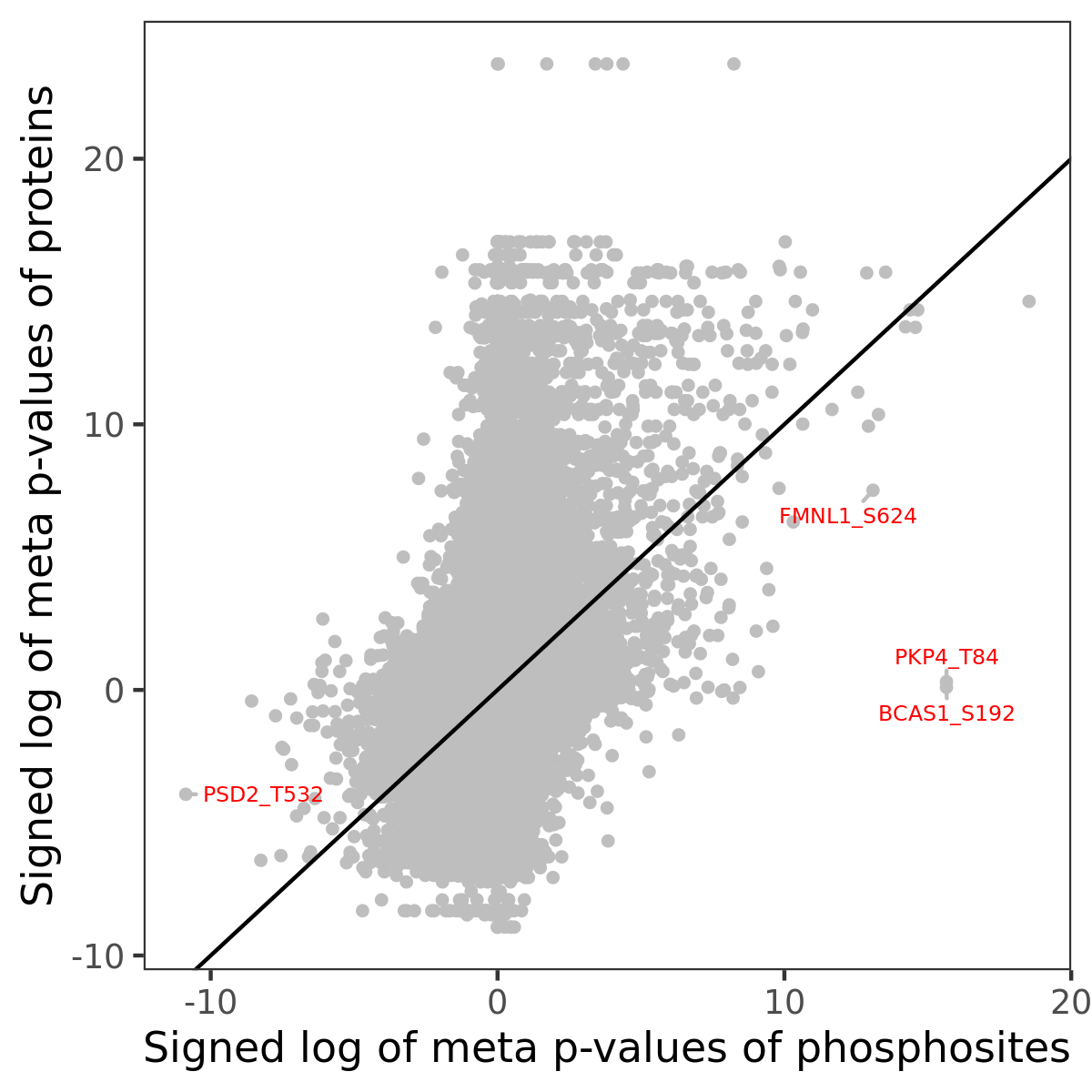
| Signed p-values | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Protein | Site | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC |
Gene set enrichment analysis
Submit phosphorylation sites and the common logarithm of the p-values of their association with xcell: Mast cell to WebGestalt.