Basic information
- Phenotype
- xcell: Granulocyte-monocyte progenitor
- Description
- Enrichment score inferring the proportion of granulocyte-monocyte progenitors in the tumor derived from RNA data using the xCell tool.
- Source
- https://genomebiology.biomedcentral.com/articles/10.1186/s13059-017-1349-1
- Method
- The R package immunedeconv (V2.0.4) (PMID: 31510660) was used to perform immune cell deconvolution using RNA expression data (TPM).
- Genes
-
- ANAPC5
- ANP32A
- AP2M1
- APLNR
- APPL1
- ARHGAP6
- ARID1A
- ARPP21
- ATXN10
- B3GNT4
- BMS1
- CALCOCO2
- CBX3
- CCNT2
- CD33
- CD5L
- CDH9
- CDK5RAP1
- CEACAM4
- CEACAM8
- CEP63
- CFP
- CHD4
- CHD9
- CLC
- CLDN17
- CLEC1B
- CLP1
- CNOT8
- COG4
- COMMD8
- COX7A2L
- CPA3
- CPSF7
- CRHBP
- CRYGD
- CSF1R
- CSPP1
- CTSG
- CXCL9
- CXorf21
- DDOST
- DDX21
- DDX27
- DEFA4
- DEK
- DHX9
- DNAJA2
- DNAJC2
- DNAJC7
- DNMT1
- DNTT
- DUSP12
- EIF3A
- EIF4E
- ELANE
- ENO1
- ENOSF1
- ERCC3
- ERP29
- EWSR1
- FEN1
- FLT3
- FOXI1
- G6PC3
- GABPA
- GAR1
- GAS8
- GGCX
- GLMN
- GLRA2
- GNL2
- GOLGA7
- GPATCH8
- GPN1
- GPR3
- GSTM5
- GTPBP4
- GUCY2D
- HDAC3
- HDC
- HEATR1
- HMGB2
- HMGXB4
- HNRNPA1
- HNRNPA2B1
- HNRNPA3
- HNRNPD
- HNRNPH3
- HNRNPM
- HNRNPR
- HPGDS
- HPS4
- IK
- IL3RA
- IL3RA_PAR_Y
- IMPDH2
- INSL3
- IPO5
- IQCC
- IQSEC3
- IRGQ
- ITGA9
- KCNJ14
- KIF17
- KPNA2
- LAMB4
- LAS1L
- LDHB
- LPAR4
- LPO
- LUC7L3
- LUZP1
- MAP7D3
- MC4R
- MED9
- MEFV
- METAP1
- MPO
- MRPL20
- MRPL3
- MRPL9
- MS4A3
- MTIF2
- MUL1
- MYOZ3
- NCL
- NCOA4
- NCOR1
- NDUFC2
- NGLY1
- NIT2
- NOL7
- NOP16
- NPM1
- NR2C1
- OMD
- PAFAH1B2
- PARK7
- PEX2
- PNN
- POLE3
- POU2F1
- PPIP5K2
- PRG2
- PRKG2
- PRPF40A
- PRRC1
- PRTN3
- PSMA4
- PUM1
- RAG2
- RBM19
- RBM25
- RBM39
- RBM4B
- RFC1
- RNASE2
- RNASE3
- RNF220
- RPL15
- RPL5
- RPRD2
- RWDD1
- RYR3
- SELP
- SERPINB10
- SERPINI2
- SETD4
- SF3B2
- SLC24A1
- SLC25A14
- SLC25A36
- SLC5A5
- SLN
- SMC5
- SMG7
- SMNDC1
- SRP9
- SSB
- STIL
- STXBP3
- SUPV3L1
- SUV39H1
- TAF15
- TCP1
- TCTN2
- TEC
- THRAP3
- TOP2B
- TRH
- TRIP11
- TTC26
- TTC27
- TTF2
- TXNL4B
- UBA2
- UBA3
- UBB
- UBE2G2
- UFC1
- UMPS
- USP36
- VPREB1
- VPS54
- WDR12
- WDR41
- WDR43
- WDR60
- XPO1
- ZBTB39
- ZC3H15
- ZNF207
- ZNF35
- ZNF593
- ZNF710
- More...
Gene association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in bar plots. The scatter plot highlights significant associations diven by protein rather than mRNA abundance.
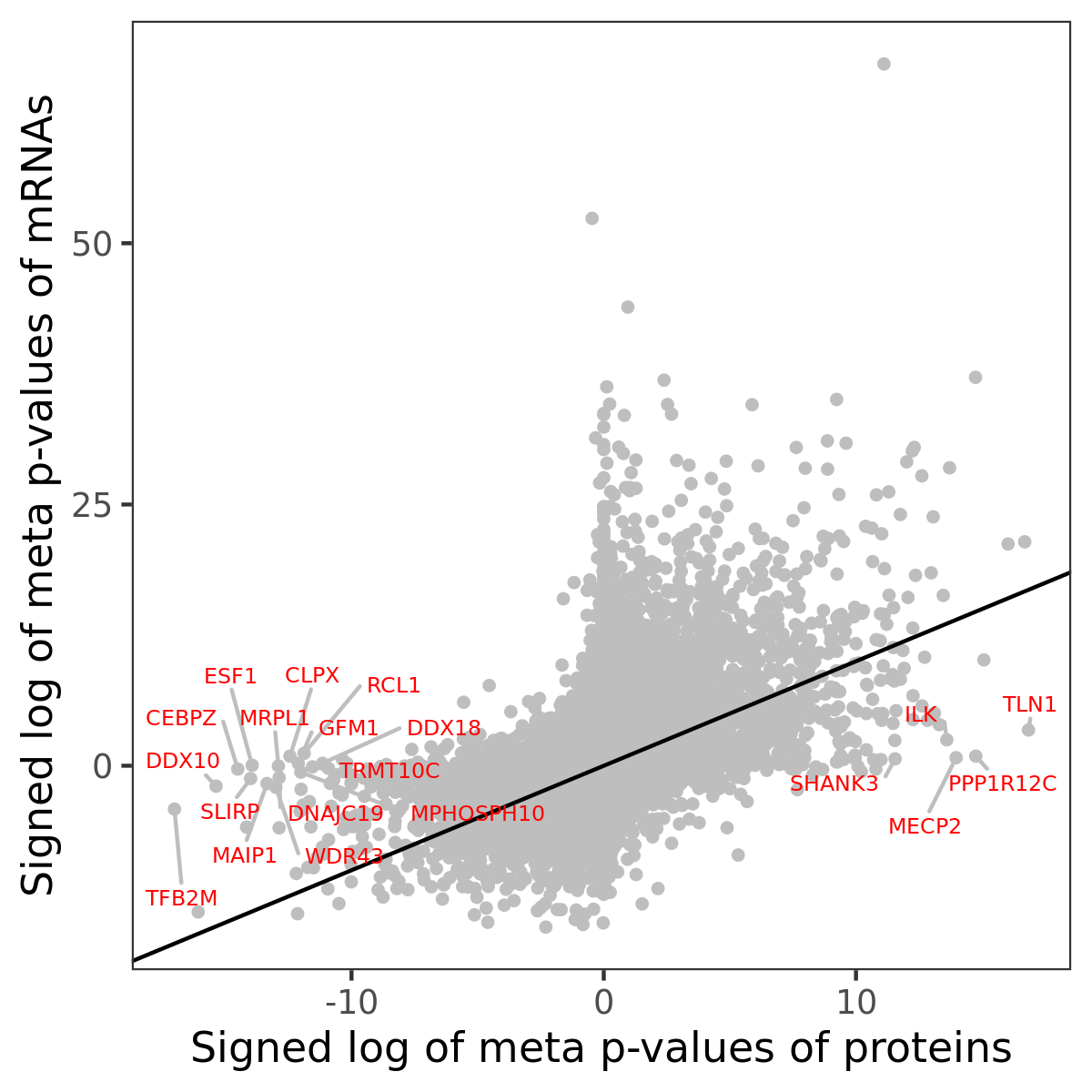
Association of protein abundance of genes
| Signed p-values | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC |
Gene set enrichment analysis
Submit genes and the common logarithm of the p-values of their association with xcell: Granulocyte-monocyte progenitor to WebGestalt.
Phosphosite association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in the bar plot. The scatter plot highlights significant associations diven by phosphorylation rather than protein abundance.
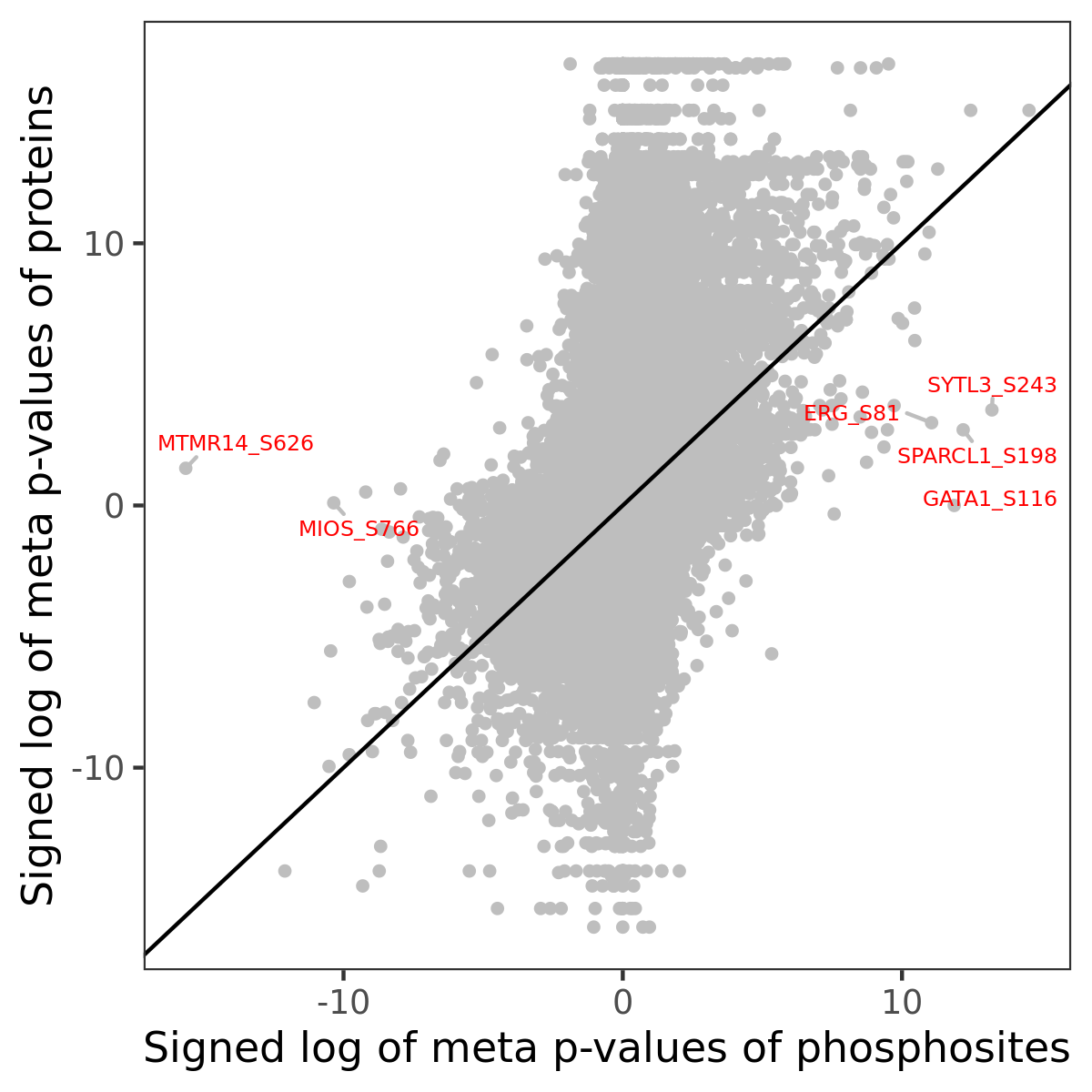
| Signed p-values | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Protein | Site | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC |
Gene set enrichment analysis
Submit phosphorylation sites and the common logarithm of the p-values of their association with xcell: Granulocyte-monocyte progenitor to WebGestalt.