Basic information
- Phenotype
- HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION
- Description
- Enrichment score representing genes involved in the epithelial to mesenchymal transition. The score was calculated using ssGSEA on the RNA data (Z-score transformed coding genes with expression in at least 50% of the samples in a cohort).
- Source
- http://www.gsea-msigdb.org/gsea/msigdb/cards/HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION.html
- Method
- Single sample gene set enrichment analysis (ssGSEA) was performed for each cancer type using gene-wise Z-scores of the RNA expression data (RSEM) for the MSigDB Hallmark gene sets v7.0 (PMID: 26771021) via the ssGSEA2.0 R package (PMID: 30563849). RNA data were filtered to coding genes with < 50% 0 expression. (Parameters: sample.norm.type="rank", weight=0.75, statistic="area.under.RES", nperm=1000, min.overlap=10). Pathway activity scores are normalized enrichment scores from ssGSEA.
- Genes
-
- ACTA2
- ADAM12
- ANPEP
- APLP1
- AREG
- BASP1
- BDNF
- BGN
- BMP1
- CADM1
- CALD1
- CALU
- CAP2
- CAPG
- CCN1
- CCN2
- CD44
- CD59
- CDH11
- CDH2
- CDH6
- COL11A1
- COL12A1
- COL16A1
- COL1A1
- COL1A2
- COL3A1
- COL4A1
- COL4A2
- COL5A1
- COL5A2
- COL5A3
- COL6A2
- COL6A3
- COL7A1
- COL8A2
- COLGALT1
- COMP
- COPA
- CRLF1
- CTHRC1
- CXCL1
- CXCL12
- CXCL6
- CXCL8
- DAB2
- DCN
- DKK1
- DPYSL3
- DST
- ECM1
- ECM2
- EDIL3
- EFEMP2
- ELN
- EMP3
- ENO2
- FAP
- FAS
- FBLN1
- FBLN2
- FBLN5
- FBN1
- FBN2
- FERMT2
- FGF2
- FLNA
- FMOD
- FN1
- FOXC2
- FSTL1
- FSTL3
- FUCA1
- FZD8
- GADD45A
- GADD45B
- GAS1
- GEM
- GJA1
- GLIPR1
- GPC1
- GPX7
- GREM1
- HTRA1
- ID2
- IGFBP2
- IGFBP3
- IGFBP4
- IL15
- IL32
- IL6
- INHBA
- ITGA2
- ITGA5
- ITGAV
- ITGB1
- ITGB3
- ITGB5
- JUN
- LAMA1
- LAMA2
- LAMA3
- LAMC1
- LAMC2
- LGALS1
- LOX
- LOXL1
- LOXL2
- LRP1
- LRRC15
- LUM
- MAGEE1
- MATN2
- MATN3
- MCM7
- MEST
- MFAP5
- MGP
- MMP1
- MMP14
- MMP2
- MMP3
- MSX1
- MXRA5
- MYL9
- MYLK
- NID2
- NNMT
- NOTCH2
- NT5E
- NTM
- OXTR
- P3H1
- PCOLCE
- PCOLCE2
- PDGFRB
- PDLIM4
- PFN2
- PLAUR
- PLOD1
- PLOD2
- PLOD3
- PMEPA1
- PMP22
- POSTN
- PPIB
- PRRX1
- PRSS2
- PTHLH
- PTX3
- PVR
- QSOX1
- RGS4
- RHOB
- SAT1
- SCG2
- SDC1
- SDC4
- SERPINE1
- SERPINE2
- SERPINH1
- SFRP1
- SFRP4
- SGCB
- SGCD
- SGCG
- SLC6A8
- SLIT2
- SLIT3
- SNAI2
- SNTB1
- SPARC
- SPOCK1
- SPP1
- TAGLN
- TFPI2
- TGFB1
- TGFBI
- TGFBR3
- TGM2
- THBS1
- THBS2
- THY1
- TIMP1
- TIMP3
- TNC
- TNFAIP3
- TNFRSF11B
- TNFRSF12A
- TPM1
- TPM2
- TPM4
- VCAM1
- VCAN
- VEGFA
- VEGFC
- VIM
- WIPF1
- WNT5A
- More...
Gene association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in bar plots. The scatter plot highlights significant associations diven by protein rather than mRNA abundance.
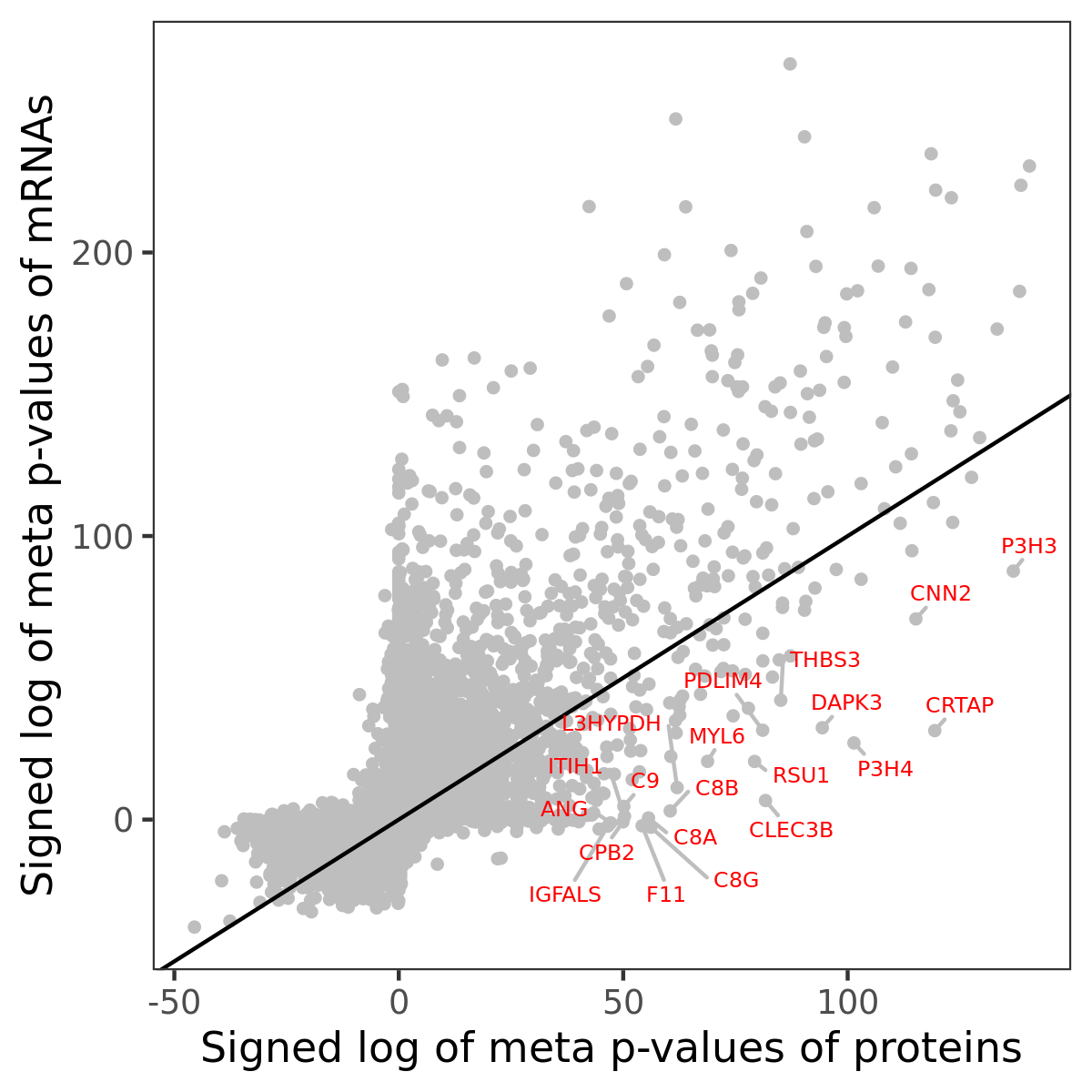
Association of protein abundance of genes
| Signed p-values | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC |
Gene set enrichment analysis
Submit genes and the common logarithm of the p-values of their association with HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION to WebGestalt.
Phosphosite association
Number of significant genes with P-value ≤ 10-6 for each cohorts are summarized in the bar plot. The scatter plot highlights significant associations diven by phosphorylation rather than protein abundance.
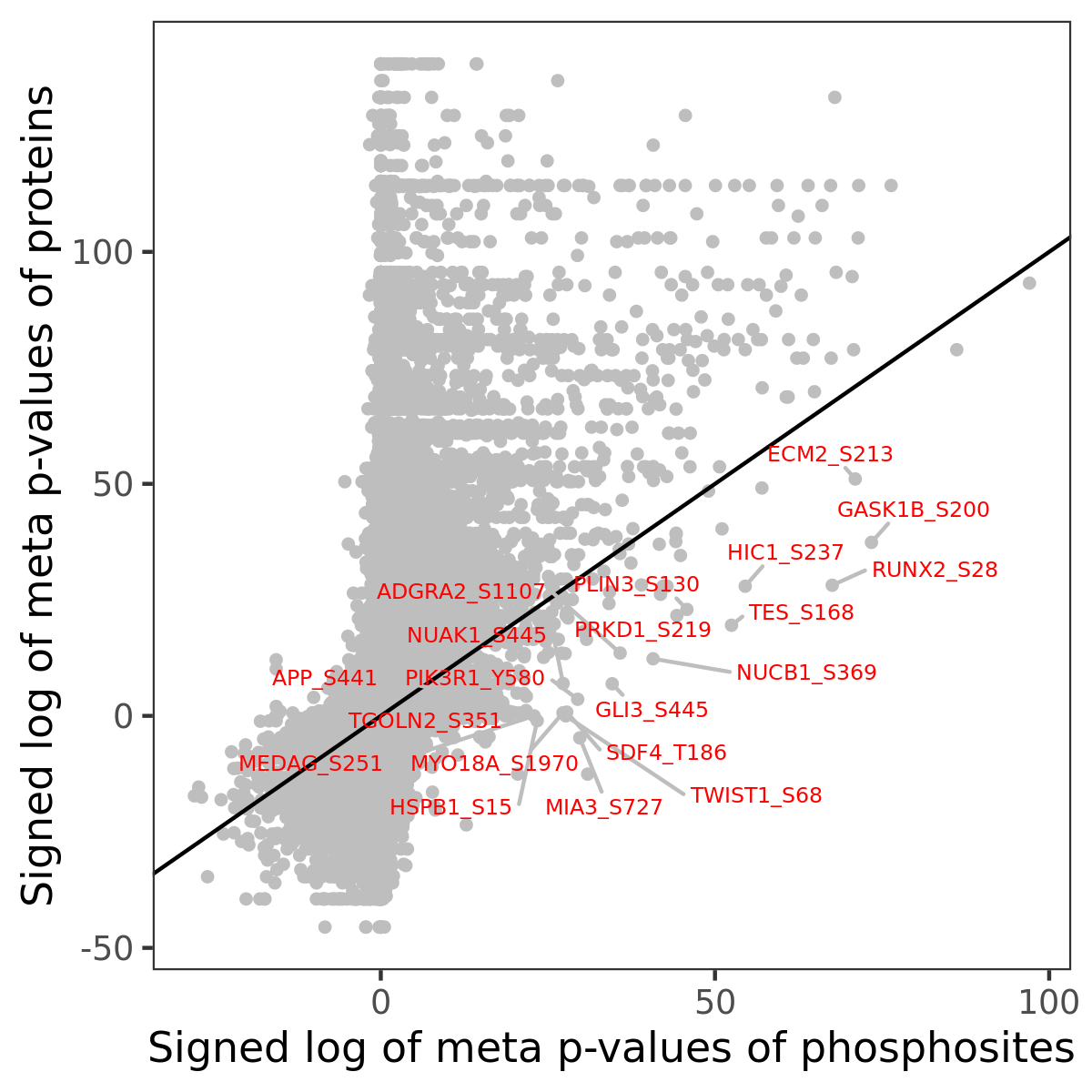
| Signed p-values | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Protein | Site | Meta P | BRCA | CCRCC | COAD | GBM | HNSCC | LSCC | LUAD | OV | PDAC | UCEC |
Gene set enrichment analysis
Submit phosphorylation sites and the common logarithm of the p-values of their association with HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION to WebGestalt.